R for reproducible scientific analysis
Data frames and reading in data
Learning Objectives
- Read tabular data from a file into a program.
- Select individual values and subsections from data.
- Display simple graphs.
We are studying the changes in total population, life expectancy and gross national income per capita (US $ by exchange rate) for 142 countries over a period of 55 years with measurements taken every 5 year, from 1952 to 2007. The data sets are stored in comma-separated values (CSV) format. Each row holds the observations for a country at one time point. The columns hold the following information: country, year, total population, continent, life expectancy and gross national income per capita. For more information visit the Gapminder website.
The first few rows of our file look like this:
country,year,pop,continent,lifeExp,gdpPercap
Afghanistan,1952,8425333,Asia,28.801,779.4453145
Afghanistan,1957,9240934,Asia,30.332,820.8530296
Afghanistan,1962,10267083,Asia,31.997,853.10071
Afghanistan,1967,11537966,Asia,34.02,836.1971382
Afghanistan,1972,13079460,Asia,36.088,739.9811058
Afghanistan,1977,14880372,Asia,38.438,786.11336
Afghanistan,1982,12881816,Asia,39.854,978.0114388
Afghanistan,1987,13867957,Asia,40.822,852.3959448
Afghanistan,1992,16317921,Asia,41.674,649.3413952
We want to:
- Load data into memory,
- Find the country with the highest gross national income per capita, and
- Plot the total population of Argentina over the years.
Loading Data
To load our gapminder data, first we need to tell our computer where is the file that contains the values. We know its name is gapminder-FiveYearData.csv. This is very important in R, if we forget this step we’ll get an error message when trying to read. We can change the current working directory using the function setwd. For this example, we change the path to the working directory where our project is stored that is named test_project:
setwd("~/test_project")Alternatively you can change the working directory using the RStudio GUI using the menu option Session -> Set Working Directory -> Choose Directory...
We also know that experimental files are located in the directory data inside the working directory. Now we can load the data into R using read.csv:
read.csv(file = "data/gapminder-FiveYearData.csv", header = TRUE)The expression read.csv(...) is a function call that asks R to run the function read.csv.
read.csv has two arguments: the name of the file we want to read, and whether the first line of the file contains names for the columns of data. The filename needs to be a character string (or string for short), so we put it in quotes. Assigning the second argument, header, to be TRUE indicates that the data file has column headers.
Since we didn’t assign the output of read.csv to any variable, the console will display the full contents of the file gapminder-FiveYearData.csv onto the interactive R console.
Challenge 1
Go to File -> New File -> R Script, and write a R script to read the gapminder dataset and assign it to a variable named gapminder .
Save the R script as data_analysis under a new folder in your project named scripts/ and add it to version control.
Run the script using the source function, using the file path as its argument (or by pressing the “source” button in RStudio).
What is the data structure of gapminder? (hint: use function class)
Data frames are similar to matrices, except each column can be a different atomic type. Underneath the hood, data frames are really lists, where each element is an atomic vector, with the added restriction that they’re all the same length. Data frames are very useful for storing data and you will find them elsewhere when programming in R. A typical data frame of experimental data contains individual observations in rows and variables in columns.
We can see the dimensions, or shape, of the data frame with the function dim:
dim(gapminder)[1] 1704 6
This tells us that our data frame, dat, has 1704 rows and 6 columns. Alternatively, you can see the number of rows and columns using the functions nrow and ncol, respectively.
Slicing data using indices
If we want to get a single value from the data frame, we can provide an index in square brackets, just as we do in math:
# first value in dat
gapminder[1, 1][1] Afghanistan
142 Levels: Afghanistan Albania Algeria Angola Argentina Australia ... Zimbabwe
R automatically stored this first column as a factor, not a character vector. We can change this by coercing the column to character vector:
gapminder$country <- as.character(gapminder$country)
class(gapminder$country)[1] "character"
An index like [1, 1] selects a single element of a data frame, but we can select whole sections as well. For example, we can select the first five measurements (rows) like this:
gapminder[1:5, ] country year pop continent lifeExp gdpPercap
1 Afghanistan 1952 8425333 Asia 28.801 779.4453
2 Afghanistan 1957 9240934 Asia 30.332 820.8530
3 Afghanistan 1962 10267083 Asia 31.997 853.1007
4 Afghanistan 1967 11537966 Asia 34.020 836.1971
5 Afghanistan 1972 13079460 Asia 36.088 739.9811The slice 1:5 means, “Start at index 1 and go to index 5”. To select all the columns you simply don’t include a slice for the those and R returns them all. If we don’t provide a slice for either rows or columns, e.g. dat[, ], R returns the full data frame.
The slice does not need to start at 1, e.g. the line below selects rows 5 through 10:
gapminder[5:10, ]We can use the function c to select non-contiguous values using both indices(for the rows) and names (for the columns):
gapminder[c(3, 8, 37, 56), c("country", "year", "pop")] country year pop
3 Afghanistan 1962 10267083
8 Afghanistan 1987 13867957
37 Angola 1952 4232095
56 Argentina 1987 31620918
Conditional slicing of data
To extract all the measurements for Argentina, we use conditional slicing:
gapminder[gapminder$country == "Argentina", ] country year pop continent lifeExp gdpPercap
49 Argentina 1952 17876956 Americas 62.485 5911.315
50 Argentina 1957 19610538 Americas 64.399 6856.856
51 Argentina 1962 21283783 Americas 65.142 7133.166
52 Argentina 1967 22934225 Americas 65.634 8052.953
53 Argentina 1972 24779799 Americas 67.065 9443.039
54 Argentina 1977 26983828 Americas 68.481 10079.027
55 Argentina 1982 29341374 Americas 69.942 8997.897
56 Argentina 1987 31620918 Americas 70.774 9139.671
57 Argentina 1992 33958947 Americas 71.868 9308.419
58 Argentina 1997 36203463 Americas 73.275 10967.282
59 Argentina 2002 38331121 Americas 74.340 8797.641
60 Argentina 2007 40301927 Americas 75.320 12779.380
The condition operator is applied to every element of country column of the gapminder data frame, only to return those rows of gapminder for which the country is “Argentina”.
Challenge 2
Find which country(-ies) had the highest gdpPercap throughout all years.
Discuss with your neighbors. Have you come up with the same answer? Have you > used the same command(s)?
Plotting
The mathematician Richard Hamming once said, “The purpose of computing is insight, not numbers,” and the best way to develop insight is often to visualise data. Visualisation deserves an entire lecture (or course) of its own, but we can explore a few of R’s plotting features.
Let’s take a look at the population of Argentina over time. Plotting the values is done with the function plot.
dat <- gapminder[gapminder$country=="Argentina", ]
plot(dat$year, dat$pop)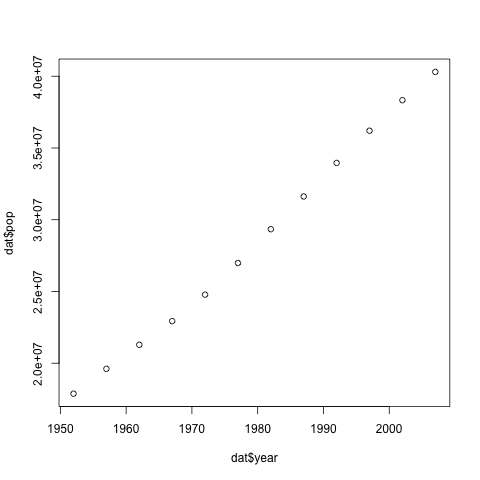
Above, we gave the function plot a vector of numbers corresponding to the population of Argentina over the years and the years. plot created a scatter plot where the y-axis is the population and the x-axis are the years.
